Screening of a soil metatranscriptomic library by functional complementation of Saccharomyces cerevisiae mutants.
Introduction
Aim of this study was to detect functional transcripts in a soil metatranscriptomic library to complement yeast mutants, here his3- and pho5-. Using this “functional” approach we are able to produce and characterize important fungal enzymes, which are potentially involved in soil/biogeochemical processes (i.e. phosphatase in P cycle).
Methods
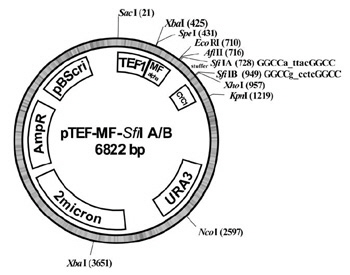
We used extracted RNA from an organic horizon of a forest soil. cDNA was synthezised using SMART technology (Clontech), the amplified cDNA contained SfiI A/B sites on the transcript ends to allow a directional ligation into a modified yeast expression vector (pTEF-MF-SfiI A/B, see picture).
The vector was first transformed in E. coli, and positiv transformants were pooled (~13,000). From this pool, vectors including the soil inserts were extracted (big plasmid prep) and used for transformation of yeast mutants.
Results and Discussion
Four candidates complementing the pho5- mutation were found.
The candidates were sequences and one phosphatase gene (acid phosphatase/phytase), probably belonging to a soil basidiomycete was characterized.
The full annotated gene:
Literature
Kellner H., Luis P., Portetelle D., Vandenbol M. (2011): Screening of a soil metatranscriptomic library by functional complementation of Saccharomyces cerevisiae mutants. Microbiological Research 166: 360-368