|
6. Increased atmospheric N deposition alters fungal cellobiohydrolase and laccase expression.
study main contributer: Ivan Edwards, Michigan; Prof. Zaks group
Abstract PLoS One:
High levels of atmospheric nitrogen (N) deposition may result in greater terrestrial carbon (C) storage. In a northern hardwood ecosystem, exposure to over a decade of simulated N deposition increased C storage in soil by slowing litter decay rates, rather than increasing detrital inputs. To understand the mechanisms underlying this response, we focused on the saprotrophic fungal community residing in the forest floor and employed molecular genetic approaches to determine if the slower decomposition rates resulted from down-regulation of the transcription of key lignocellulolytic genes, by a change in fungal community composition, or by a combination of the two mechanisms. Our results indicate that across four Acer-dominated forest stands spanning a 500-km transect, community-scale expression of the cellulolytic gene cbhI under elevated N deposition did not differ significantly from that under ambient levels of N deposition. In contrast, expression of the ligninolytic gene lcc was significantly down-regulated by a factor of 2–4 fold relative to its expression under ambient N deposition. Fungal community composition was examined at the most southerly of the four sites, in which consistently lower levels of cbhI and lcc gene expression were observed over a two-year period. We recovered 19 basidiomycete and 28 ascomycete rDNA 28S operational taxonomic units; Athelia, Sistotrema, Ceratobasidium and Ceratosebacina taxa dominated the basidiomycete assemblage, and Leotiomycetes dominated the ascomycetes. Simulated N deposition increased the proportion of basidiomycete sequences recovered from forest floor, whereas the proportion of ascomycetes in the community was significantly lower under elevated N deposition. Our results suggest that chronic atmospheric N deposition may lower decomposition rates through a combination of reduced expression of ligninolytic genes such as lcc, and compositional changes in the fungal community.
After consistently finding lower phenoloxidase activity in soils under increased N deposition (i.e. Michigan Gradient Study; Sugar Maple forests), we conducted this study to find the molecular microbial mechanisms behind it. Thus we measured the enzyme activity in the whole gradient using MUB-cellobioside for cellobiohydrolases and ABTS for laccase and found except for one site a constantly decrease in enzyme activities.
Consequently we analysed the expression of fungal laccase genes and cellobiohydrolase (GH family 7) genes with quantitative PCR and found a significant downregulation of the fungal laccase genes, but not the cellobiohydrolase genes.
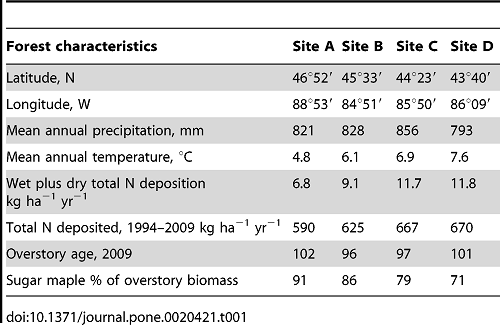 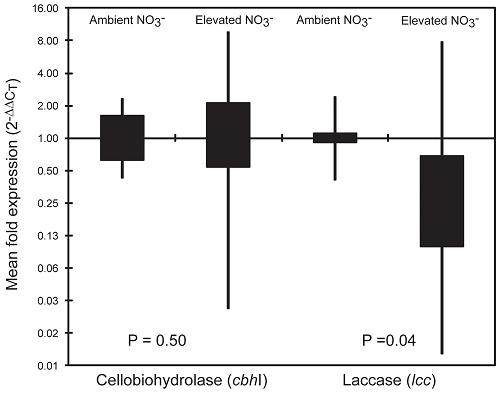
Table 1. Site, climatic, overstory and ambient nitrogen deposition rates of four sugar maple stands receiving experimental NO3- additions.
Figure 1. Relative expression of fungal cbhI and lcc genes in the forest floor of northern hardwood forests under ambient or elevated levels of N deposition. Boxes define the interquartile range, whiskers mark the minimum and maximum observations; relative expression (Y-axis) is log-scaled. P-values are significance of the mean difference in relative expression under ambient and elevated N deposition (Mann-Whitney U, n = 21).
Table 2. Cellobiohydrolase and laccase enzyme activities and gene expression levels under ambient and elevated NO3− deposition in four northern hardwood forests.
Another question, but not tested to a full extent, was to know if the fungal community might change following an increased N deposition. This we followed only for one site. Nevertheless, there are indications that the active community (“expressed 28S rRNA gene”) is shifting. Moreover it gave us a glimpse what fungal species are active in this study site.
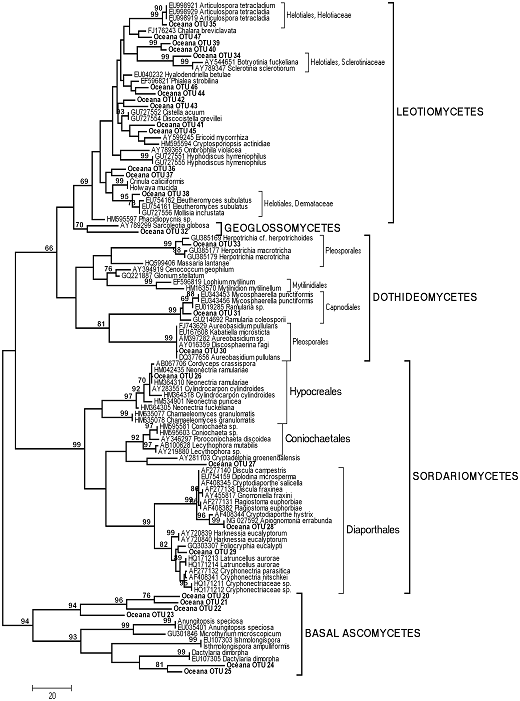 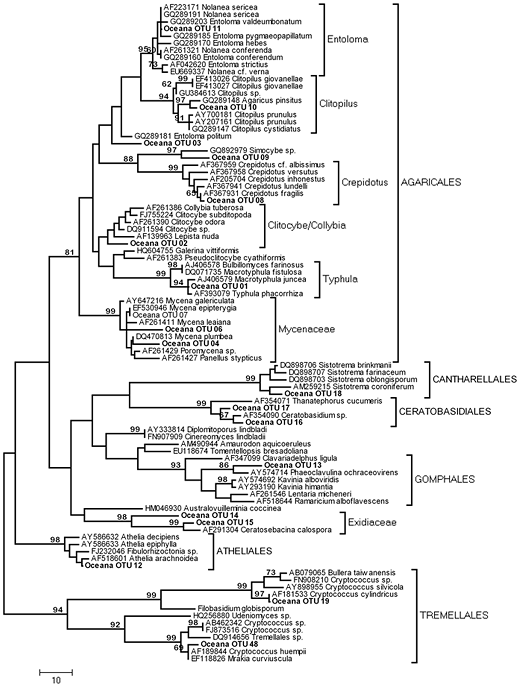
Figure 2. Phylogenetic relationships between 28 environmental Ascomycete sequences recovered from a maple-dominated hardwood site (Site D, “Oceana”) and 79 representative Ascomycete sequences recovered from GenBank. Tree represents the 50% consensus of 43 most parsimonious trees (tree length 1391) inferred from ca. 500 bp at the 5′ end of the nuclear large subunit. MP bootstrap values >65% are shown above nodes.
Figure 3. Phylogenetic relationships between 19 environmental Basidiomycete sequences recovered from a maple-dominated hardwood site (Site D, “Oceana”) and 73 representative Basidiomycete sequences recovered from GenBank. Tree represents the 50% consensus of 33 most parsimonious trees (tree length 885) inferred from ca. 500 bp at the 5′ end of the nuclear large subunit. MP bootstrap values >65% are shown above nodes.
Nevertheless we found a remarable diversity, and could also verify that Mycena species might play (again) an important role in forest ecosystems. The shift of the community was calculated by a CCA.
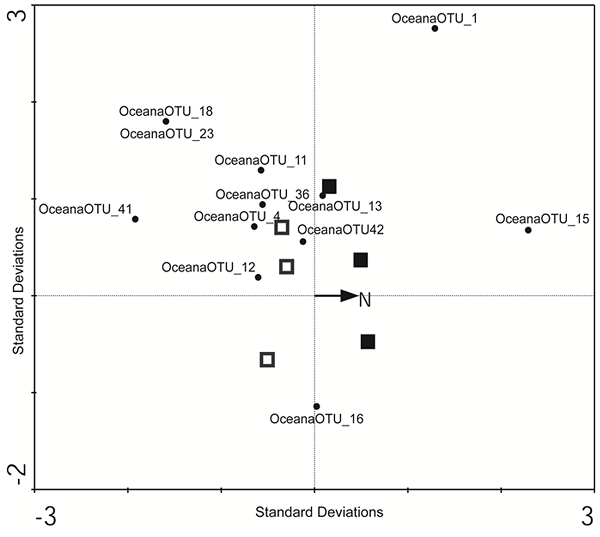 |
Figure 4. Triplot based on a Canonical Correspondence Analysis showing the relationships between samples of a fungal community developing under ambient conditions of N deposition (open squares) and under conditions of simulated elevated N depostion (closed squares), the relative abundance of the 12 most widespread fungal Operational Taxonomic Units (OTUs, black circles), and total N depostion (vector) in the forest floor of a maple dominated hardwood ecosystem. The primary axis accounts for 22% of the variance in OTU relative abundances; the second axis a further 37%. The relationship between fungal relative abundances and total N is not significant (Monte Carlo P = 0.52). OTU codes correspond to the phylogenies (Fig. 2, Fig. 3): OceanaOTU_1, Typhula sp; OceanaOTU_4, Mycena sp; OceanaOTU_11, Entoloma sp; OceanaOTU_12, Athelia sp.; OceanaOTU_13, Gomphales sp.; OceanaOTU_15, Ceratosebacina sp.; OceanaOTU_16, Ceratobasidium sp.; OceanaOTU_18, Sistotrema sp.; OceanaOTU_23, unidentified Ascomycota; OceanaOTU_36, Leotiomycete sp.; OceanaOTU_41, Leotiomycete sp.; OceanaOTU_42, Leotiomycete sp
Reference
Edwards I.P., Zak D.R., Kellner H., Eisenlord S.D., Pregitzer K.S. (2011): Simulated atmospheric N deposition alters fungal community composition and suppresses ligninolytic gene expression in a northern hardwood forest. PLoS ONE 6(6): e20421.
doi:10.1371/journal.pone.0020421
|