|
3. Development of a proteomic approach to detect and identify fungal laccases
second project phase of SPP1090 - done by Nico Jehmlich, Dirk Benndorf & me
Aim of this study was to detect the laccase proteins, first in pure culture systems. This might enable in future studies to detect them too in soil systems etc. Dirk Benndorf developed the peptides for immunisation and production of the antisera in silico - using a multiple alignment of fungal laccase proteins. The advantage was to take a conserved copper binding region (cbr II) of the laccases, which made it possible to use the antibody with different laccases of different fungi (particularly basidiomycetes).
But, based on the conserved position of the cbr in the core of the protein, it was necessary to denature the laccase proteins to make a binding of the antibody possible. The test´s for functionality and sensitivity presented here were done by Nico and me.
A first step was to compare the functionality of the antibody LccCbr2 with native and denatured fungal laccases (we used Hypholoma fasciculare and Coprinopsis cinerea laccase, see Fig. 1.).
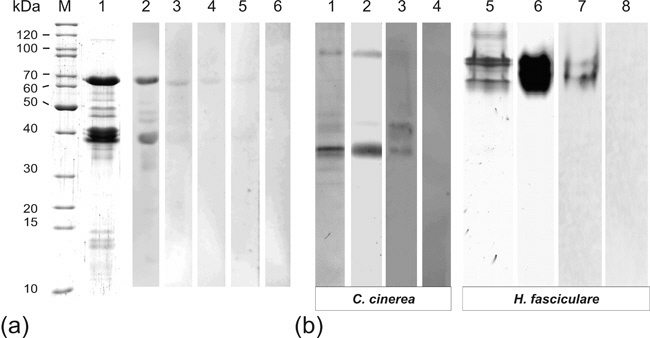 |
Fig. 1. Characterization and functionality of the LccCbr2 antibody. (a) Inhibitory antibody incubation. Five micrograms of the highly purified Coprinopsis cinerea laccase Lcc1 were stained with Coomassie Brilliant Blue (CBB) (lane 1) or incubated and detected together with the antibody LccCbr2 (lane 2) and with 10 nmol (lane 3), 50 nmol (lane 4), 125 nmol (lane 5) and 150 nmol (lane 6) of the originally synthesized four peptides together with the antibody LccCbr2, respectively. Protein marker (PageRulerTM Unstained Protein Ladder, Fermentas, Ontario, Canada) is indicated in lane M. (b) Separation of extracellular laccase from C. cinerea and Hypholoma fasciculare cell-free culture supernatants using native-PAGE without mass rulers. The extracellular proteins of the supernatants were functional stained using 1mM ABTS (lanes 2 and 6) and total proteins were stained using CBB (lanes 1 and 5). Laccase was immunologically detected by chemiluminescence (2 min exposure). The gel was incubated in 2% SDS-solution for 15 min prior to Western blotting and incubation with LccCbr2 antibody (lanes 3 and 7). No signal appeared for C. cinerea and H. fasciculare when proteins were not denatured prior to Western blotting and incubation with LccCbr2 antibody (lanes 4 and 8).
Our finding was, that the antibody was only working with denatured laccase proteins. The LccCbr2 cannot match to the epitope in the inner nativ core of the laccase, where the cbr2 is located. Thus we used in a second step denatured laccases of several basidiomycetes to check if we could detect them with the antibody (Fig. 2.).
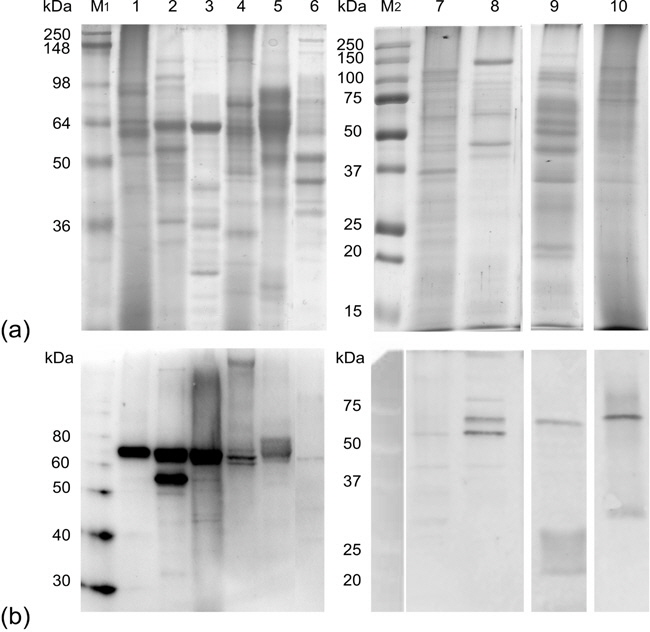 |
Fig. 2. (a) SDS-PAGE of extracellular fungal laccases concentrated from cultural supernatants. (b) Detection after Western blotting and antibody incubation using chemiluminescence (2 min exposure). A total of 75g protein was loaded per lane. Lane 1: Hypholoma fasciculare, 2: Trametes versicolor, 3: Coprinopsis cinerea, 4: Pleurotus cornucopiae, 5: Mycena sp., 6: Piriformospora indica, 7: Verpa conica, 8: Pleurotus ostreatus, 9: Pycnoporus cinnabarinus and 10: Lentinula edodes. Protein markers are indicated in lanes M (a: M1= SeeBlue Plus2 Pre-Stained Standard (Invitrogen), M2= Precision Plus ProteinTM Prestained Standard (Bio-Rad, Hercules, USA); b: M1= MagicMarkTM XP Western Protein Standard (Invitrogen), M2= Precision Plus ProteinTM Prestained Standard (Bio-Rad) detected under white light).
As a result we were able to detect strong bands via chemiluminescence detection from several basidiomycetes (MW ~ 70 kDa). These protein bands were cut from the SDS gel, tryptic digested and measured via mass spectrometry (peptide fingerprintig). So far we were only able to detect the laccases from 5 species by MS (Table 1).
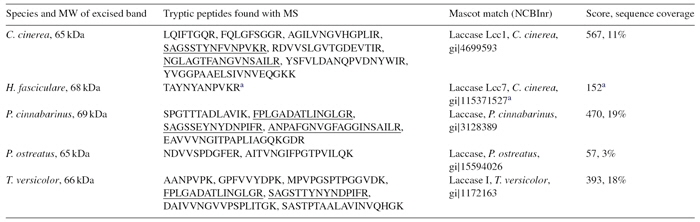 |
Table 1. Identification of fungal laccase proteins via analysis of tryptic peptides by mass spectrometry. Peptides matching the same regions in more than one species are underlined.
a The peptide was de-novo sequenced using data from tandem mass spectrometry by the software Spectrum Mill (Agilent Technologies, Paolo Alto, California). The score in the table represents the Sherenga score of the sequenced peptide
However, this might be a first step to get laccases from environmental samples. Moreover the laccase antibody is now also running in other lab groups to detect more laccases.
Kellner H., Jehmlich N., Benndorf D., Hoffmann R., Rühl M., Hoegger P.J., Majcherczyk A., Kües U., von Bergen M., Buscot F. (2007): Detection, quantification and identification of fungal extracellular laccases using polyclonal antibody and mass spectrometry. Enzymes and Microbial Technology 41: 694-701. doi:10.1016/j.enzmictec.2007.06.002-701.
|