|
2. Temporal fungal laccase gene diversity and expression in a forest Cambisol
second project phase of SPP1090
The aim of this study was to analyze the changes of fungal laccase gene diversity and expression in the most diverse organic horizon of a forest soil within one year. Moreover these changes were also related to the soil laccase activity. To keep site effects to a minimum we decided to sample 3 plots (20 x 20 cm) within an homogenous Fagus sylvatica stand without ground vegetation. DNA and RNA were extracted from 3 homogenized subsamples per plot and per sampling date. Enzyme activity was measured in 20 samples per sampling date around the 3 plots. Altogether 6 sampling dates were analysed within one year. All amplified laccase genes were recorded and related to fungal reference sequences (especially references obtained from fruitbodies of this particular site). We used the well working primer combination Cu1F & Cu2R, as well as the new combination Cu1AF & Cu2R aiming to amplify ascomycete laccase genes. In each plot, 25 clones were analysed.
2.1. Dominance structure
A total of 450 clones were analyzed using amplification products of the primer combination Cu1F & Cu2R, yielding in 408 basidiomycete laccase genes. After NJ analysis, 73 distinct laccase genes were differentiated. The new primer combination Cu1AF & Cu2R did not amplify ascomycete laccase, but instead bacterial laccase genes (see Section IV; this happens because the cbr’s are very conserved among different organisms!). Note: however, the combination works perfectly with soil cDNAs and amplified reasonable amounts of expressed ascomycete laccase genes (see below!).
All different laccase gene types were recorded in each analyzed plots, which then gave the characteristic dominance structure (Fig. 1). We characterized 14 types to be dominant as we found them independently in min. 5 soil samples (plots). As shown in previous studies, the most dominant types could be related to different Mycena and Russulaceae species (several at 100% identity level). Interestingly, 11 of 74 types could we 100% relate to known reference sequences that we amlified from fruitbodies of this particular site.
The laccase types were related to single species, genera or families and their probable status of nutrition (saprotrophic or mycorrhizal), which was important for the following analyses. A remarkable result so far was, that only 5 types which we amplified from soil DNA could be related to expressed sequences.
Which gives the important question: how to handle this “bias”?
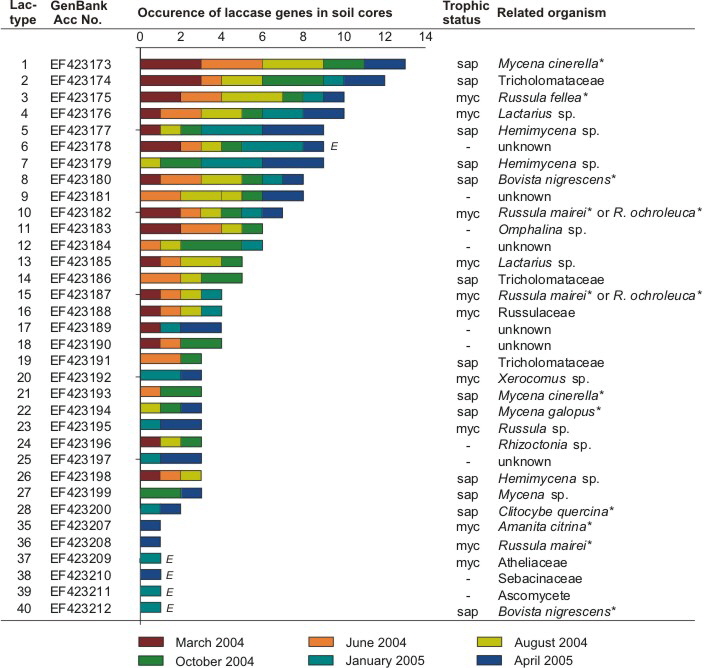 |
Fig. 1: Dominance structure of environmental basidiomycete laccase gene types found with a minimum appearance of three times independently. Laccase genes found as singletons and doubletons were only shown if they had identical fungal references or found to be expressed (E). Accordingly GenBank accession numbers, sampling date, sampling frequency (one to three soil cores), relation to laccase genes from fungal reference strains and their possible trophic status (myc – mycorrhizal, sap – saprotrophic) are given. Species marked with * were collected with fruiting bodies at the forest site. Laccase gene types marked with “E” were also found expressed.
2.2. Variation in presence of the basidiomycete laccase genes in soil samples
We found a remarkable variation in the presence of basidiomycete laccase genes over time. There was an increase in laccase gene types, with a peak in October, then a decrease (Fig. 2). We counted also the abundance of each type (up to 3, for the 3 plots), and the increase was even more dramatic (see publication). However, we belive that especially the rare saprotrophic and rare unknown types account most for this increase (for example the rare Clitocybe quercina ... etc).
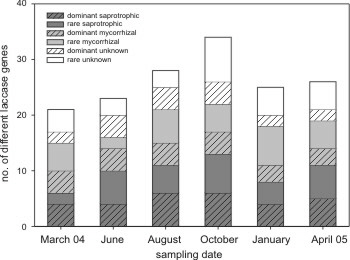 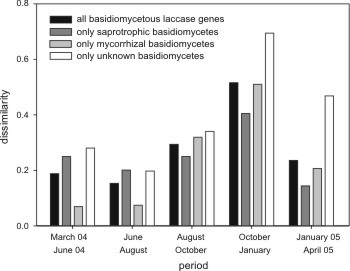
Fig. 2 (left): Number of different unique laccase gene types (excluding frequence) per sampling date, found in all 3 plots. The nutritional status is determined according NJ-tree to known fungal references (and their known ecology) - separating in saprotrophic, mycorrhizal and unknown ecologies. Rare types are found less than 5 times in different soil samples.
Fig. 3 (right): Dissimilarity of the pooled laccase gene community (pooled data of 3 plots) between consecutive samplings.
We compared the change of the population of consecutive samplings and found the highest dissimilarity from October 2004 to January 2005 (Fig. 3). Probably a lot of rare fungi appear or disappear in this time period.
2.3. Variation in the expression and relation to soil laccase activity
The analysis of 180 expressed laccase gene sequences, which were obtained with both primer pairs from soil RNA extracts (cDNA), revealed 42 different laccase gene types. Noteworthy, only five expressed laccase genes corresponded to genes amplified from the soil DNA extracts. Highest expressions (qualitative & quantitative) were found over the period August, October and January (Fig. 4). However, these considerable changes in the expression pattern were not reflected in the overall soil laccase activity.
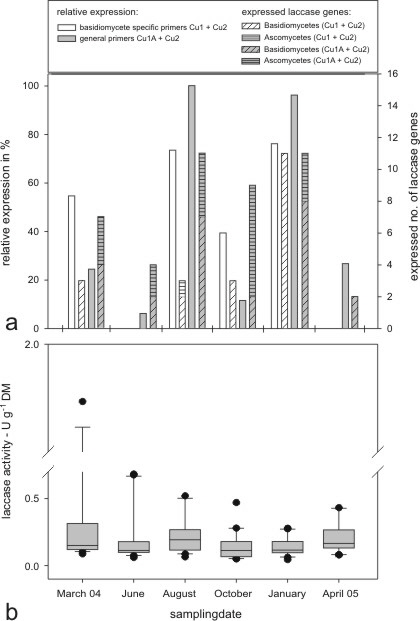 |
Fig. 4: Characterization of the expression of fungal laccase genes found with the two different primer sets, giving gene richness and quantification of the PCR product (a). The found genes were identified according cluster analysis and divided in asco- and basidiomycetes. The total expressed gene richness (S) is given on the top line and the number of expressed genes of ascomycetes is given in brackets. Activity of extracellular laccases in the soil samples (n=20) is given for each sampling date (b). Median values are represented as central line within the 95% grey shaped box.
2.4. Conclusion
Using the momentarily available molecular tools, we found distinct present (genomic DNA) and expressed laccase populations (see publication), questioning which approach to use in future research. Analyzing of genomic populations proved useful when comparing different forest ecosystems, soil horizons or ecosystem manipulations. However, genomic soil derived populations seem not to represent the active, expressed populations. Future increases in databases (GenBank) might partially resolve this problem.
We could demonstrate distinct temporal changes of the laccase gene community composition and expression that were not reflected by variations in the total activity of extracellular laccases. Abiotic factors like temperature or rainfall did not correlate to observed population richness or enzyme activities (data not shown). However, we found that the changes in the type and diversity level of present and expressed laccase genes might be a response to temporal fluctuations in the differentiation of soil micro-niches, triggered by annual inputs of leaf litter. Such micro-niches are possibly more structured during and shortly after incorporation of fresh litter into the O-horizon, leading to an enhanced diversity in presence and expression of laccase genes to maintain a similar enzyme activity as in phases with lowered niche differentiation.
Remarkably is the strong fungal activity in the winter (it was relatively mild). However, there are several indications that fungal activity might dominating in the winter time and bacterial activity during the spring/summer.
2.5. The reliability of the laccase NJ-tree
Several questions arise how to relate the soil sequences to references. We use different measures to relate the unknown sequences to fungal references. First, you can use the normal phylogenetic tools and analysis of the exons, which unfortunately only include about 100 nucleotide positions between cbr I & II. Second, some intron positions are remarkedly conserved among fungal taxa or genera (e.g. Mycena species). And third, it is important to gain reference strains or DNA from fruitbodies of the investigated site. We found a considerable amount of identical hits between our soil DNA sequences and laccase sequences from sampled fruitbodies.
However, this short exon will make it more and more difficult to analyse the unknown sequences of soil samples in future attempts. We have indications, that there are also distinct mixed subfamilies appearing within the larger laccase phylogeny. In future studies it might be usefull to amplify larger laccase gene regions, if possible (and make use of aminoacid alignments).
 |
Fig. 5: Schematic representation of a typical fungal laccase between copper binding regions cbr I and cbr II. Numbers beneath show the different found introns and their insertion position. Boxes marked with “ins” and “del” indicates positions, where amino acid insertion or deletions were found. (attention: the consensus for cbr II is GTFWYHSH)
Table 1: Characterization of the intron occurrence in all analyzed LMCO sequences, giving position, length, reading frame and fungi including it.
|
Intron no. (see Fig.1)
|
No´s. of LMCO gene types including it
|
Insertion position
|
Length of intron (bp)
|
Frame
|
Fungal references including it
|
|
1
|
1
|
26
|
71
|
+1
|
Macrotyphula juncea
|
|
2
|
11
|
45
|
49-55
|
+2
|
Mycena cinerella, M. crocata, M. galopus, M. zephirus
|
|
3
|
2
|
51
|
51-53
|
+2
|
Pleurotus ostreatus
|
|
4
|
1
|
53
|
51
|
+1
|
Botryotinia fuckeliana
|
|
5
|
6
|
62
|
51-55
|
+1
|
Mycena zephirus
|
|
6
|
6
|
64
|
53-60
|
0
|
Rhizoctonia solani
|
|
7
|
2
|
65
|
52
|
+1
|
Macrotyphula juncea, Mycena rosea
|
|
8
|
1
|
80
|
61
|
+1
|
unknown
|
|
9
|
1
|
81
|
49
|
+2
|
unknown
|
|
10
|
1
|
92
|
53
|
+1
|
Mycena rosea
|
|
11
|
5
|
109
|
52-58
|
0
|
Mycena crocata, M. galopus
|
|
12
|
10
|
118
|
53-61
|
0
|
Bovista nigrescens, Hemimycena crispata, Piloderma byssinum, P. crocata, Tylospora fibrillosa
|
|
13
|
44
|
121
|
52-63
|
0
|
Hemimycena crispata, nearly all Russula sp., all Lactarius sp., Trametes sp.*, Phlebia sp.*, Lentinula sp.*, Ganoderma sp.*, Nematoloma sp.**, Clitocybula sp.**
|
|
no intron
|
77
|
-
|
-
|
-
|
nearly all Ascomycota, many Basidomycota (exceptions see above)
|
|
* see D’Souza et al. (1996), ** see Scheel et al. (1999) - these references had no homologous soil sequences and are not part of the NJ tree!
Note: the laccase genes 16 & 17 of Coprinopsis cinerea (Hoegger et al., 2006) include multiple introns between cbr I & II and are not included in the figure and table. The evolutionary history and the biological usage of these genes remain at the moment unclear.
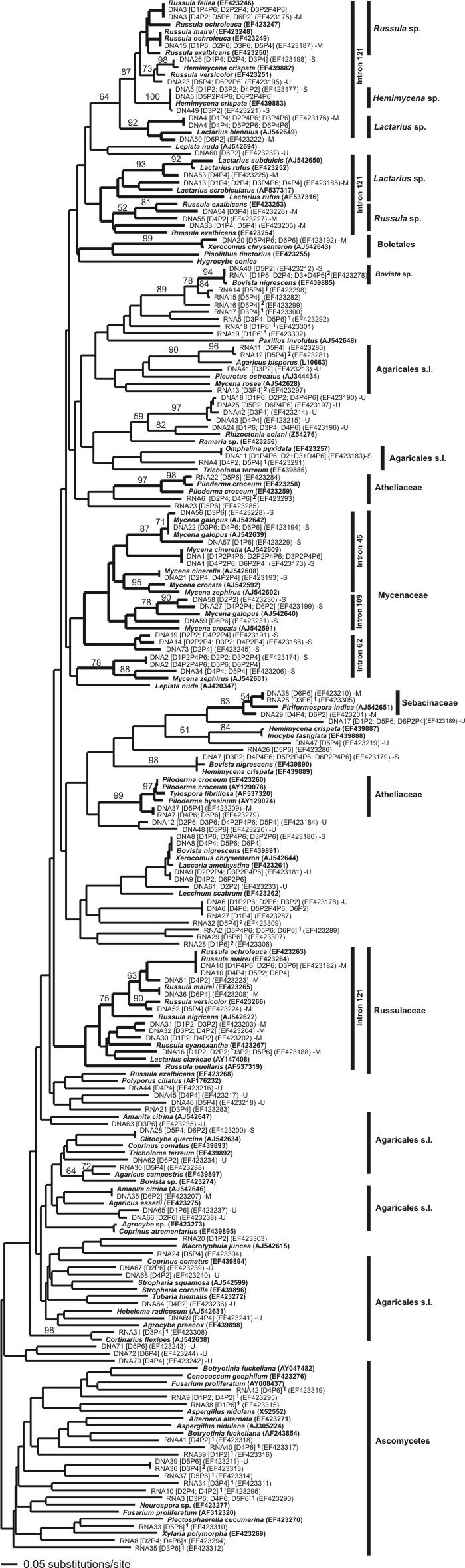 |
Fig. 6. Neighbor-joining tree calculated from the coding region of fungal laccase genes obtained from soil samples as well as fungal references using K2P-distances. Branch support was assessed by 2,000 bootstrap replicates, which are given above branches. Occurrence of introns supporting fungal taxa is indicated. Major monophyletic fungal groups are indicated with bold branches, their trophic status is indicated (m, mycorrhizal; s, saprotrophic; u, unknown). The occurrence of each environmental laccase gene sequence is given in brackets (D1 - D6 indicating sampling dates Mar 04 – Apr 05; sampling plots: P2, P4, P6) with their accordingly GenBank accession number. Expressed laccase genes, high ranked “1” refers to a sequence obtained with primer combination Cu1Af/Cu2r, whereas “2” indicates the amplification with both primer sets and without number means the finding only with primer set Cu1f/Cu2r.
The laccase gene alignment used for this study is freely available [here] or at treebase (study no.: S2321)
References
D’Souza, T.M., Boominathan, K. & Reddy, C.A. (1996) Isolation of laccase gene-specific sequences from white rot and brown rot fungi by PCR. Appl Environ Microbiol 62: 3739-3744.
Hoegger, P.J., Kilaru, S., James, T.Y., Thacker, J.R., Kües, U., (2006) Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS Journal 273: 2308-2326.
Kellner H., Luis P., Zimdars B., Kiesel B., Buscot F. : Diversity of bacterial laccase-like multicopper oxidase genes in forest and grassland Cambisol soil samples. Soil Biol Biochem 41: 1380-1389.
doi:10.1016/j.soilbio.2009.03.012
Scheel, T., Hölker, U., Ludwig, S., Höfer, M., (1999) Evidence for and expression of a laccase gene in three basidiomycetes degrading humic acids. Appl Microbiol Biotechnol 52: 66-69.
Errata
100-year-old mixed stand of European beech (F. sylvatica L.) and Pedunculate oak (Quercus robur L.) is not correct in the publication. It is a mixed European beech (F. sylvatica L.) and Sessile oak (Quercus petraea) stand.
“metric potential” in the introduction should read “matric potential”
|