|
Detection of expressed fungal type I polyketide synthase genes in a forest soil
1. Research questions
Information on the PKS gene diversity in and among soil microbial communities is described for actinobacteria, but we do not understand the complete community of soil microorganisms possessing these genes, let alone whether they are expressed in the soil environment. In this initial study, we focused on the presence of fungal (i.e. ascomycete) reducing PKS (HR-PKS) genes and their expression in a forest soil, to provide evidence for the importance of this gene in soil processes.
2. Methods of investigation
Soil sampling was performed in a sugar maple dominated forest soil in Oceana County, MI [link]. RNA and DNA were extracted from the homogenized soil samples (organic horizon, each ~1 g). cDNAs were synthesized from extracted RNA according SMART technology (Clonetech). The method we employed to synthesize cDNA produces full-length gene transcripts, because adapter sequences at both ends of the transcripts allow amplification of all cDNAs by a long-distance PCR (SMART technology, Clonetech). Soil DNA was separated from RNA using the RNA/DNA Midi kit (10) loaded on agarose gel and purified using the QIAquick Gel Extraction Kit (Qiagen). Partial fungal type I polyketide synthase genes, spanning part of the ketoacyl synthase (KS) and acyl transferase (AT) domain, were amplified using the KAF1 (GAR KSI CAY GGI ACI GGI AC) and KAR2 (CCA YTG IGC ICC YTG ICC IGT RAA) primer pair (Amnuaykanjanasin et al. 2005). PCR products (~ 600-950 bp) were gel-purified using the QIAquick Gel Extraction Kit (Qiagen) and cloned into the pCR 2.1-TOPO vector using the TOPO TA Cloning kit (Invitrogen, Carlsbad, USA). A total of 20 clones of each PCR reaction were randomly selected, cultured overnight in liquid Luria broth, and sequenced using standard M13 primers (GenBank accession numbers: FJ232770-FJ232791).
3. Results and perspectives
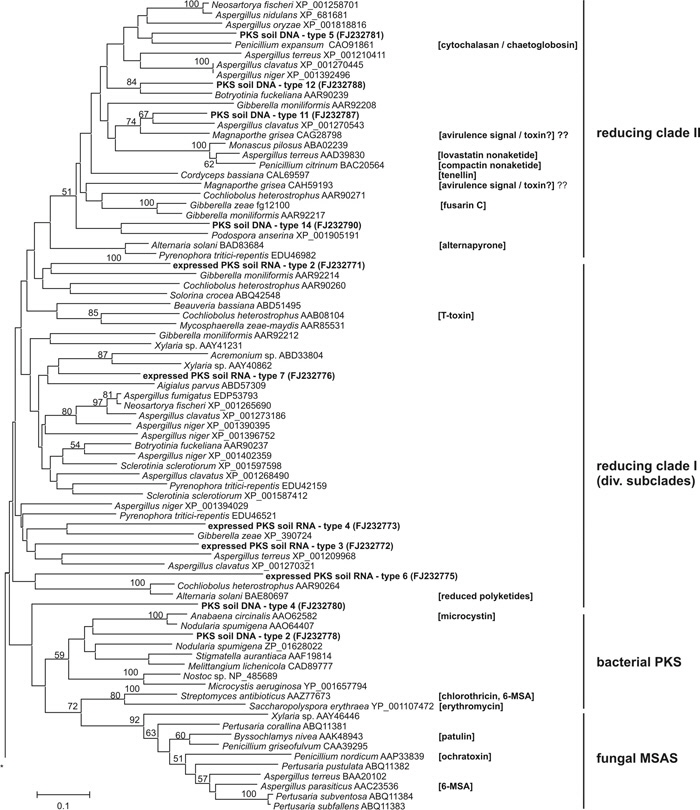
We amplified, sequenced and identified 15 PKS genes present in forest floor DNA (Fig. 2). The length of the amplified partial genes ranged from 663-941 bp, and some sequences contained one (PKS soil DNA - type 9 & 15, Fig. 1) or two putative introns (PKS soil DNA - type 1 & 14). The newly obtained genes clustered in previously defined clades (reducing clades I-IV, Kroken et al. 2003). Seven different expressed reducing PKS genes could be amplified and sequenced from the cDNA banks, ranging in length from 603-813 bp (Fig. 2). Surprisingly, none of these sequences matched PKS genes that we amplified from forest floor DNA. The PKS genes amplified from cDNA clustered in two defined clades (reducing clades I and III). PKS RNA-type 1 was the most frequent, and it was present in two of our three cDNA banks, whereas all other types appeared exclusively in one library (i.e., singletons).
In addition to the mere presence of fungal type I PKS genes in soil, we could clearly demonstrate their expression in the soil environment. This observation suggests that soil ascomycetes deploy this metabolic pathway under field conditions, and the products of the PKS pathway may be used to enhance their competitive ability in soil. Hence, interactions for niche competition and substrate acquisition (succession of leaf litter in autumn by ascomycetes) seem to ecologically explain our findings. However, future studies need to analyze temporal and spatial differences in the expression and identify produced metabolites, which would provide further insight into their importance for microbial community dynamics in soil, especially between ascomycete fungi and other organisms. If these expressed genes, i.e. PKSs produce compounds that elicit antagonistic interactions among soil microorganisms, then the production of PKSs may be an important factor controlling the composition of soil microbial communities.
 |
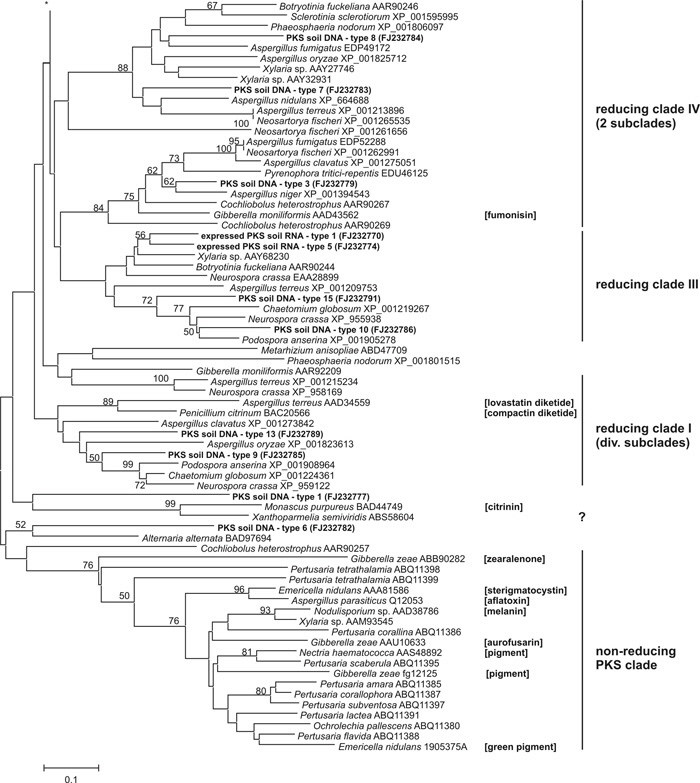 |
Fig. 2: Phylogenetic tree of partial PKS amino acid sequences using Neighbor-Joining method and Poisson corrected distances. Branch support was estimated by calculating 2,000 bootstrap replicates and is shown above the branches. Additional information for product formation of single PKS genes is reported in brackets, clade structure information is based on Kroken et al. (2003). Discarding hypervariable alignment proportions, effectively 71 positions were used to generate the phylogenetic inference. PKS genes Gibberella zeae fg12100 and fg12125 are obtained from the munich information center for protein sequences (mips.gsf.de/genre/proj/fusarium).
The amino acid aligment of the amplified genes can be downloaded here, or in TreeBASE (study no. S2290) (http://www.treebase.org/treebase/index.html)
4. References
Kellner H., Zak D.R., 2009. Detection of expressed fungal type I polyketide synthase genes in a forest soil. Soil Biology & Biochemistry 41: 1344-1347.
doi:10.1016/j.soilbio.2009.02.033
Amnuaykanjanasin, A., Punya, J., Paungmoung, P., Rungrod, A., Tachaleat, A., Pongpattanakitshote, S., Cheevadhanarak, S., Tanticharoen, M., 2005. Diversity of type I polyketide synthase genes in the wood-decay fungus Xylaria sp. BCC 1067. FEMS Microbiology Letters 251:125-136.
Kroken, S., N. L. Glass, J. W. Taylor, O. C. Yoder, and B. G. Turgeon. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. U.S.A. 100:15670-15675.
Schümann, J., and C. Hertweck. 2006. Advances in cloning, functional analysis and heterologous expression of fungal polyketide synthase genes. J. Biotechnol. 124:690-703.
Erratum
I wrote in the publication about cDNA libraries. This is not correct, it are cDNAs containing adaptor sequences, which made it suitable to reamplify the whole cDNA whenever it is necessary. But per definition a cDNA library are all single cDNAs ligated into a vector.
|