|
1. Spatial basidiomycete laccase gene diversity and expression in a forest Cambisol
first project phase of SPP1090 - done by Patricia Luis
Standard molecular methods were used in order to investigate fungal laccases. In particular, DNA and RNA was extracted from the samples (environmental samples or collected fungal fruitbodies, RNA was reverse-transcribed into cDNA) and thereafter used in PCR for amplification of fungal laccase gene fragments. The obtained sequences were used in cluster analyses and the environmental laccase gene sequences of soil DNA or RNA (cDNA) extracts were related to sequences obtained from fungal reference straines or fruit bodies. Moreover fungal straines were screened for laccase activity using ABTS (Fig. 1).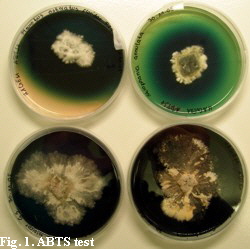
Accordingly, in a first study, Patricia Luis was able to develop degenerate primers to amplify partial laccase genes of Basidiomycetes (Luis et al., 2004). The approach was subsequently tested with DNA extracted from soil samples of a brown forest soil (Cambisol; mixed beech-oak stand; 5 soil cores devided in Oh, Ah and Bv horizon). As a result it was possible to amplify more than 60 different basidiomycete laccase genes, which could be attributed to saprotrophic, mycorrhizal and unknown fungal taxa. However, genes belonging to Mycena spp., div. Russulaceae and Atheliaceae were found to be most dominating. These results are concomitant to analyses of other researcher using ribosomal genes. Moreover it was discovered that laccase genes belonging to saprotrophic fungi seemed to be more restricted to upper horizons of this forest soil than mycorrhizal ones. However, this was expected before and was now confirmed.

The logical next step (second study) was to develop an approach to analyze the laccase gene expression (Luis et al., 2005a). The major problem was the development of the RNA extraction procedure - luckily, Patricia Luis has golden hands. Five soil cores of the organic horizon Oh were analyzed. All samples showed the expression of actin genes (housekeeping gene), whereas only two of these samples possessed a strong expression of laccase genes and the remaining only a weak or no expression. Altogether 14 different laccase genes were found to be expressed and related to reference sequences and environmental sequences. Four expressed genes (cDNA) could be related to environmental genes (DNA) of the same soil samples, but only one expressed gene could be 100% related to the fungus Lepista nuda. Despite this preliminary character, the observed differential expression might be related to soil characteristics, like rhizospheric or bulk soil.
The aim of the third study was to discover the spatial (horizontal and vertical) diversity of the laccase genes within a greater sampling campaign and to analyze their potential geographic relationship (Luis et al., 2005b; Fig. 3). Additionally, the laccase activity within all horizons was measured.
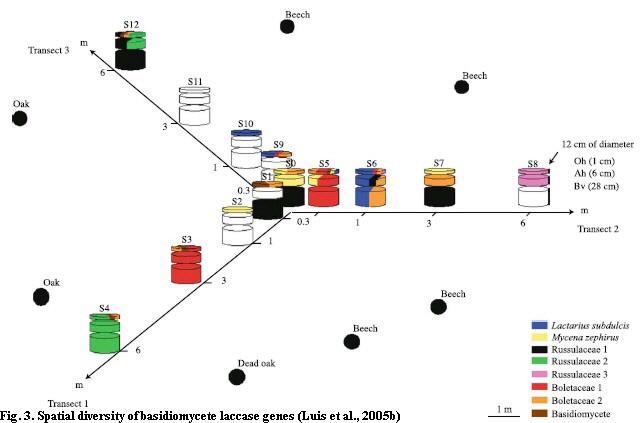 |
Altogether 167 different laccase genes (Oh: 96, Ah: 74, Bv: 50) were discovered in 13 soil cores. The diversity was higher in the organic horizon and decreased with depth. This was concomitant with the analyzed soil laccase activity (Fig. 4) and SOM content. Up to 74% of the different laccase gene sequences were found to occur only in one soil horizon, supporting previous observations of a specialized fungal community among the soil horizons. Within this study, fungal species of Russulaceae and several Mycena species dominated; the saprotrophic Mycena were frequently within the organic horizon and the mycorrhizal Russulaceae among all soil horizons of the forest Cambisol. For future sampling designs it was necessary to analyze adjacent fungal comunities. Thus, we found a dissimilarity of 67% between fungal communities of 30cm distance, which was increasing with an increasing geographical distance (Fig. 5). It simply means, if we want to analyze soil cores within a temporal approach, we might find strong dependency among replicates within approximately 20cm and independent replicates in distances of about 3 to 6 meters.
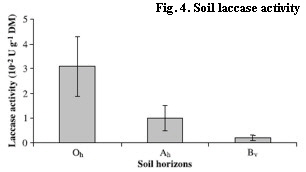 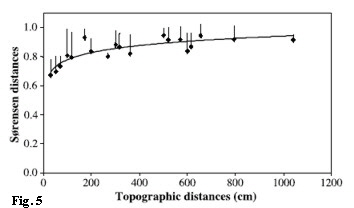
However, as we use the functional marker laccase, it was not possible to relate all found genes to known taxa. This might be a disadvantage against ribosomal markers (e.g. ITS or LSU). But using this functional gene might give clues about the potential function in soil processes, particularly if analyzing the expression. Moreover these soil functioning processes can be measured by enzyme activity tests.
The laccase gene alignment used for the third study is freely available [here] or at treebase (study no.: S1425)
Questions arising:
How does the fungal community respond temporally?
Is it possible to relate the laccase enzymes to fungal taxa?
Literature
Luis P, Walther G, Kellner H, Martin F & Buscot F (2004) Diversity of laccase genes from basidiomycetes in a forest soil. Soil Biol Biochem 36: 1025-1036. doi:10.1016/j.soilbio.2004.02.017
Luis P, Kellner H, Martin F & Buscot F (2005a) A molecular method to evaluate basidiomycete laccase gene expression in forest soils. Geoderma 128: 18-27. doi:10.1016/j.geoderma.2004.12.023P
Luis P, Kellner H, Zimdars B, Langer U, Martin F & Buscot F (2005b) Patchiness and spatial distribution of laccase genes of ectomycorrhizal, saprotrophic and unknown basidiomycetes in the upper horizons of a mixed forest Cambisol. Microb Ecol 50: 570-579.
doi:10.1007/s00248-005-5047-202
|